Introduction
Fermentation, in general, refers to the phenomenon of organic materials being catabolized by microorganisms. It is distinguished from putrefaction in that it produces compounds (or substances) that are beneficial to humans. In a broader sense, fermentation includes all phenomena in which microorganisms produce compounds (or substances) that are beneficial to humans. Therefore, “fermentation microbiology” may be described as the study of microorganisms that are useful to humankind. We continue to use the old style of Kanji (Chinese characters), “醗” of “醗酵,” which means fermentation in Japanese, in the name of our laboratory, and we hope you can feel the history of our laboratory and the potential of microorganisms in the word “醗酵学” (fermentation microbiology) of the name of our laboratory. Although the name of our laboratory may be considered old-fashioned, we are proud of this name and hope to open up new fields of fermentation microbiology in the future.
As you can see from the History of Our Laboratory, we have conducted highly original research that is ahead of its time. This tradition has been inherited and is firmly followed even now. One of our goals in the Laboratory of Fermentation Microbiology is to pursue the principle of fluidity with immutability. We believe that “immutability”—the state of remaining forever unchanged—is our approach to research:
- Believe in the potential of microorganisms proportional to their diversity.
- Consider both basic research and applied research without separating them.
- Handle a variety of subjects, such as small molecules, proteins, genes, and cells, using a variety of methods.
- Never imitate others, and conduct experiments thoroughly.
- Take pride and responsibility for your research and enjoy it.
At the Laboratory of Fermentation Microbiology, we value the research style described above.
On the other hand, the promotion of collaborative research is one of the critical aspects of “fluidity,” that is, acceptance that things should change from time to time. In recent years, the life sciences have progressed tremendously, and experimental techniques have advanced remarkably. However, we believe that fusion with other fields will be essential for future development. In the past, the Fermentation Microbiology Laboratory has placed great importance on conducting experiments “with our own hands” as much as possible. However, we now believe that development of new research fields requires active collaboration with researchers from other fields (bringing different technologies and knowledge).
We would like to explain the main research themes shown on the top page. We hope this will help you understand our research.
1. Research into the molecular mechanisms and regulatory systems behind morphological differentiation and secondary metabolism in actinomycetes
Organisms produce a variety of metabolites. Metabolites common to many organisms and essential for sustaining life—such as sugars, amino acids, lipids, and nucleic acids—are called primary metabolites. There are also unique metabolites, however—produced from primary metabolites—which are not essential for the life-sustaining processes of the organism. These metabolites are called secondary metabolites. Antibiotics and pigments are typical examples of such metabolites. Humans have used these secondary metabolites as biological resources for medicines, fragrances, and dyes for a long time.
Actinomycetes are bacteria known to produce many valuable secondary metabolites such as antibiotics. We aim to elucidate the production mechanism of these compounds, as described in Chapter 2. Another characteristic of actinomycetes is that they undergo complex morphological differentiation, growing mycelia and forming spores (Fig. 1), despite being bacteria, not fungi. For this reason, actinomycetes are important model microorganisms for basic research into cell differentiation.
For a long time, our laboratory has studied the microbial hormone A-factor, which induces morphological differentiation and secondary metabolism in Streptomyces griseus. In collaboration with other research groups, in 2008, we determined the entire genome sequence of S. griseus; we have subsequently been conducting comprehensive research, including transcriptome, ChIP/ChAP-seq, and proteome analyses. We have combined such comprehensive analyses with precise functional analysis of individual genes and proteins. The sophisticated gene expression control mechanisms and clever, flexible survival strategies of microorganisms have often surprised us, and our interest in them is endless.
Streptomyces is a major actinomycete in the soil. However, in recent years, minor soil actinomycetes have gained attention as producers of new secondary metabolites.
Since 2007, we have studied a rare actinomycete, Actinoplanes missouriensis, which forms sporangiospores on the substrate mycelium. In a moist environment, the outer membrane of the sporangium breaks, and zoospores with dozens of flagella start swimming. The zoospores swim around in the water at extremely high speed and demonstrate probable chemotaxis, moving to a suitable location for germination (Fig. 2).
(1) When vegetative mycelial growth detects particular conditions, such as nutrient depletion or dryness, sporulation (sporangium formation) begins. In this process, many genes involved in sporangium formation and spore maturation are expressed in a strictly controlled manner and sequence. By investigating the mechanisms controlling sporulation gene expression in response to environmental changes, we revealed the functions of several key regulators (see links 3, 4, 5, 6, 7). Additionally, we identified a glucosaminidase involved in spore separation in the sporangium (see link 8).
(2) The sporangium is a specialized sack-like structure that contains many dormant spores. Thus, sporangia are multicellular organs; this is very unusual among the bacteria, which are generally unicellular and reproduce by cell division. It would seem reasonable that such an unusual structure would be composed of unknown components. Therefore, our research focuses on substances that exist only in sporangia, such as the outer shell surrounding the sporangium and the matrix filling the space between spores. We are also interested in the enzymes involved in the biosynthesis and degradation of these unique substances.
(3) Spores survive in the sporangium in a dormant state, but when the surrounding environment changes from dry to wet, the outer membrane breaks, and the spores are released. We consider this dynamic change an excellent model for the process of microbial cells awakening from dormancy. We have been working to elucidate the molecular mechanism of this process to establish a new concept that has not been observed in other model organisms.
(4) Spores released from the sporangium have flagella and pili on their surface; these zoospores use their flagella to swim at very high speed through the moist environment. The zoospores show chemotaxis; in a nutrient-rich environment, they stop moving and germinate, beginning a new mycelial growth. In these processes, we are trying to elucidate some interesting phenomena such as “How do zoospores swim at such a high speed?” and “How does the movement of flagella cease when the zoospore arrives in a suitable niche?” We aim to shed light upon these fascinating phenomena. We have also clarified the role of pili in attaching to materials (see link 9).
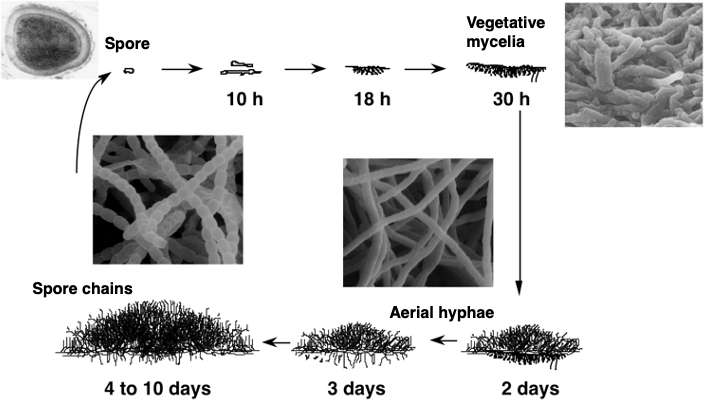
Figure 1. The life cycle of Streptomyces griseus
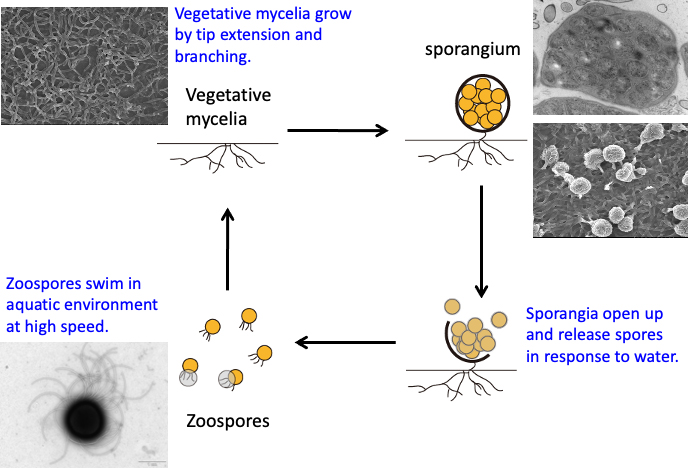
Figure 2. The life cycle of the rare actinomycete Actinoplanes missouriensis
2. Research into the biosynthesis of microbial secondary metabolites, and production of useful compounds through application of the biosynthetic machinery
Many medicines, including antibiotics, have been discovered from compounds produced (biosynthesized) by microorganisms. These compounds are usually secondary metabolites. Most secondary metabolites have unique chemical structures specific to each microbial species and are therefore highly diverse. This diversity is why so many medicines are isolated from microorganisms. The unique chemical structures of secondary metabolites are created by various biosynthetic enzymes. Thus, biosynthetic enzymes are regarded as “specialized molecular machines” in “the factory of secondary metabolites.”
With the advancement of genetic engineering technology, it has become possible to create (rather than discover) microorganisms that produce new compounds by artificially modifying various biosynthetic enzymes or using them in new combinations. This emerging technology, an important research area within synthetic biology, is gaining attention for its potential to realize environmentally friendly manufacturing processes. To use microorganisms as a “factory” for useful compounds such as new medicines and raw materials for polymers, it is crucial to prepare an extensive lineup of various biosynthetic enzymes, which are the “machines” used in the “factory.” This requires the discovery of new biosynthetic enzymes and elucidation of their functions. We are searching microbe secondary metabolism for such biosynthetic enzymes to pursue the potential of microorganisms as a “factory” of useful compounds. We are also studying biosynthetic enzymes to understand their molecular mechanisms.
2-1 Research on new biosynthetic enzymes
We use two approaches to identify new biosynthetic enzymes. One is the “compound-based” approach, which focuses on compounds with unique structures and aims to identify the novel enzymes necessary to generate these structures. In this approach, it is important to find novel compounds of interest; we are therefore actively collaborating with research groups that are working vigorously to isolate and determine the structure of microbial metabolites. Recent important results include the discovery of nitrite biosynthetic enzymes for the production of diazo groups (see link 10), diazo group synthases (see link 11), a cytochrome P450 monooxygenase that catalyzes the nitration of tyrosine (see link 12), and a cytochrome P450 that catalyzes nitrene transfer (see link 13).
The other is the “genome information-driven” approach. This is also called “genome mining.” In recent years, genome sequencing technology has progressed remarkably, revealing the genome sequences of many organisms. Within the actinomycetes alone, the genomes of thousands of species have been sequenced and made available on a database. In most cases, the genes for secondary metabolites specific to the microorganism are clustered in one place in each genome. Such a cluster of genes is called a “biosynthetic gene cluster.” Biosynthetic enzymes are regarded as “specialized molecular machines” for producing compounds with unique chemical structures. “Machines” of the same type have some similarity in their amino acid sequences. Based on these similarities, it is possible to predict the number of biosynthetic gene clusters in the genome sequence of a microorganism. Therefore, we can predict the number of secondary metabolites that are potentially produced by a microorganism whose genome sequence is known. From an actinomycete species, more than 30 different secondary metabolites are expected to be produced. Nevertheless, only two to three compounds are usually found in a single actinomycete. This means that around 90% of the metabolic capacity has been overlooked. We aim to identify new compounds and biosynthetic enzymes catalyzing unprecedented reactions by analyzing the functions of such overlooked biosynthetic gene clusters. This research has led to the discovery of new aromatic compounds (see link 14), polyene derivatives (see link 15), and siderophores (see link 16).
2-2 Elucidation of the molecular mechanisms of biosynthetic enzymes
We have explained that biosynthetic enzymes are the “machines” of secondary-metabolite production. Through recent developments in technology, research on microbial production of new useful compounds has been actively carried out by modifying or combining these “machines.” However, to modify the “machine,” it is necessary to know the molecular mechanism of the machine. Therefore, we attempt to understand the catalytic mechanisms of biosynthetic enzymes by using site-directed mutagenesis, X-ray crystallography (see link 17), and cryo-electron microscopy (see link 18).
2-3 Research on the production of valuable compounds using microorganisms
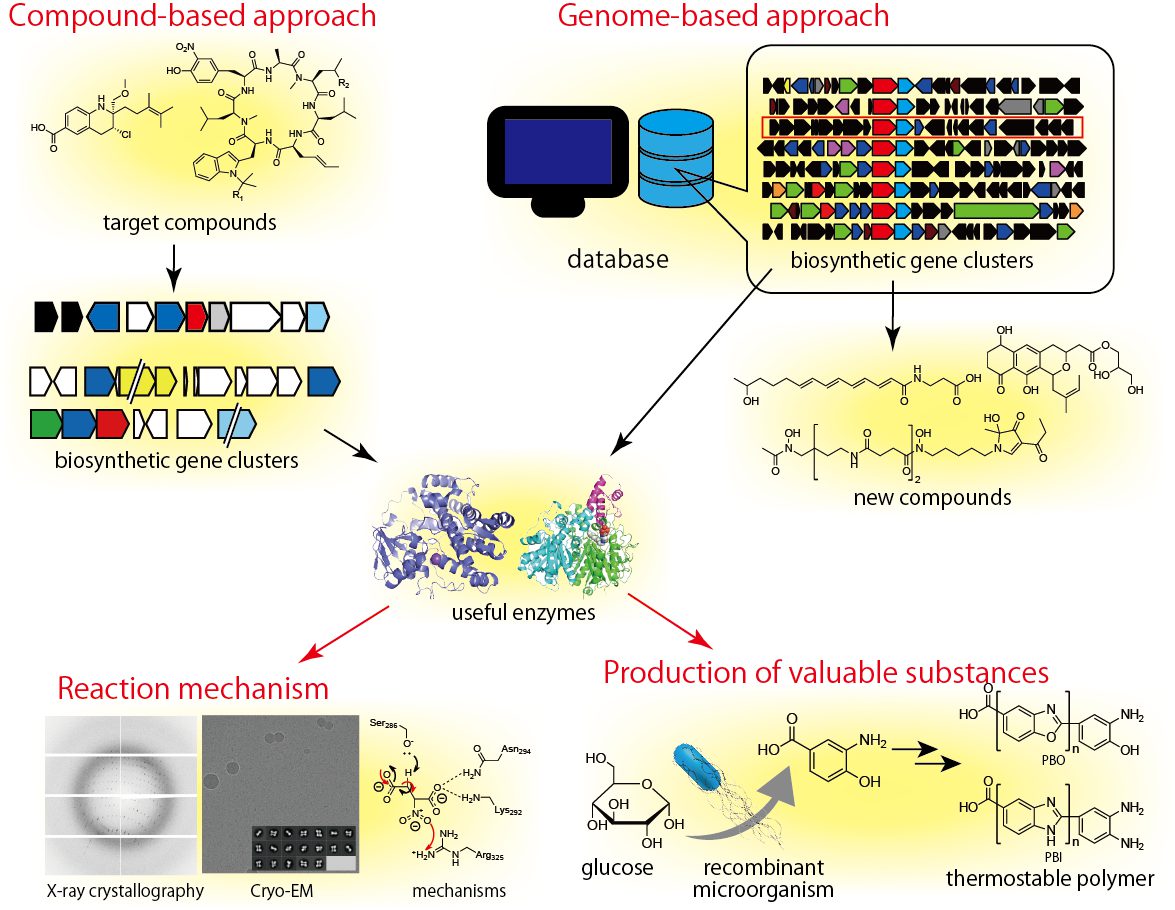
Fig. 3. Diagram of our research on the biosynthesis of secondary metabolites
